Here, P is the Projection Matrix that combines both Intrinsic and Extrinsic Parameters.
Now remember that this is homogeneous coordinates, so to get the actual pixel coordinates.
So
Because every
So, we need 6 points to solve for the 11 unknowns in P (since it's up to scale). 11 only because for eg:
Dividing by
Rearranging:
This is in the form of
To solve for P, we can use Singular Value Decomposition (SVD) on matrix A. The solution for p will be the right singular vector corresponding to the smallest singular value of A. This gives us the projection matrix P up to a scale factor.
So, with 6 points we can solve for P.
For more than 6 points:
- Overdetermined set of equations.
- We have to be careful as to avoid the trivial solution of
- If observations are noisy we are cooked, P might not exist.
NOTE! -- Cautionary Tale:
- We also don't have solution if all points are coplanar. ( i.e. Z is constant for all points )
- All points lying on a line is even worse.
- All points and projection center located on a twisted cubic curve.
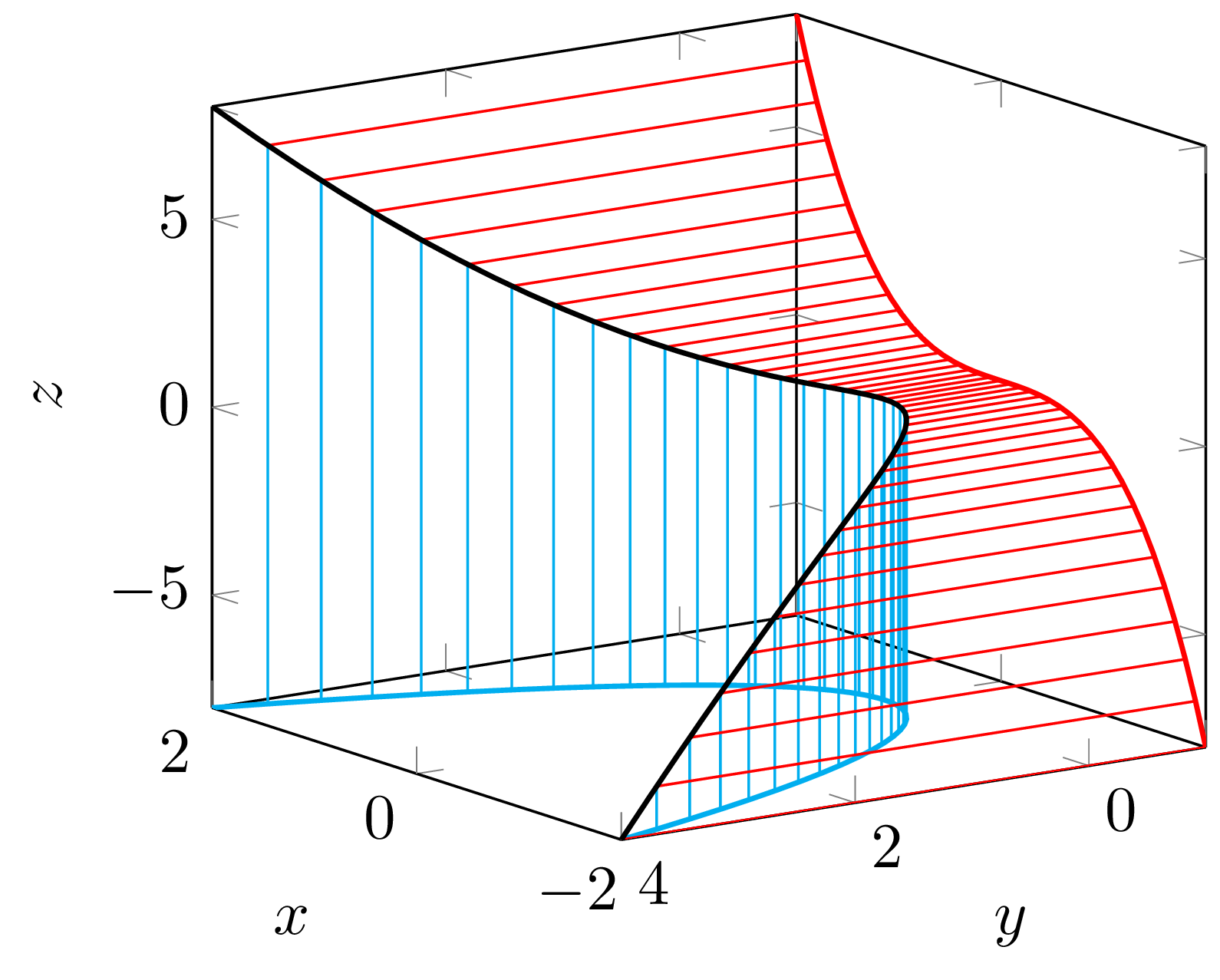
We have the system
In reality, due to noise,
If we just minimize
To fix the scale and avoid the zero vector, we add a constraint:
Now the problem is:
Minimize
using Lagrange Multipliers. Define the loss function
Take the derivative w.r.t
This is the definition of an Eigenvalue Problem:
must be an eigenvector of the matrix . is the corresponding eigenvalue.
Which eigenvector? Substitute back into the error equation:
To minimize the error, we must choose the smallest eigenvalue
Thus, the solution
The SVD Soln:
Calculating
The columns of
The singular values
Therefore, the eigenvector for the smallest eigenvalue corresponds to the smallest singular value. In standard SVD implementations, values are sorted high-to-low, so this is the last column of V (or last row of
V is 12 x 12, so the last column of V gives us the solution for P (up to scale). ( It's 12 x 12 because A is 2M x 12, so V is always 12 x 12).
After solving the DLT system
Getting K from P:
Calibration using Checkerboards ( Zhang's Method )
- Detect corners in the checkerboard images to get image points.
- The corner of the checkerboard is set to be the origin in the world frame. Also assuming that checkerboard is on the plane Z = 0 of the world frame, i.e., all points on the checkerboard have Z = 0.
So, we can remove the 3rd column of R as Z = 0 in world coordinates.
This is a homography transformation from world points on the checkerboard plane to image points.
So our long 2 x 12 matrix A reduces to a 2 x 9 matrix for each image by putting Z = 0. Which looks like:
With M points in the checkerboard, we get a 2M x 9 matrix A. We can solve for H using SVD as before.
Since H only has 8 unknowns (up to scale), we need at least 4 points (8 equations) to solve for H.
With multiple images of the checkerboard taken from different angles, we can compute multiple homography matrices
From each homography H, we can extract constraints on the intrinsic matrix K.
Extracting K from multiple Homographies
From the homography, we have:
Rearranging:
Since
Because
Because
Let
We can run Cholesky decomposition on
which follows the chol(A) = L where
Since B is symmetric, we only have to solve for 6 unique elements in B.
Defining a vector
We have
So we have to derive the equations in terms of the elements of b. Getting the G matrix such that
From the first constraint:
Expanding this gives:
This can be written in the form:
o, G becomes:
In general for i,j:
From the second equation of Homograph constraints:
So we can stack these equations for multiple images to form a matrix G such that:
With n images, we have 2n equations. To solve for the 6 unknowns in b, we need at least 3 images (6 equations).
To solve this we will do SVD on G:
Soln: Last column of V gives us b.
Which gives us our B matrix. From B we can get K using Cholesky decomposition as mentioned before.
Requirement to get K:
- At least 3 images of the checkerboard from different angles.
- At least 4 points in each image to compute Homography. ( 8 equations for 8 unknowns in H )
- Detect corners accurately in the images to get precise image points.
Original Zhang:
### Code for Zhang:
import numpy as np
def get_v_ij(H, i, j):
"""
Calculates v_ij vector (1x6) from columns i and j of Homography H.
Indices i, j are 0-based (0, 1, 2 correspond to 1, 2, 3 in math).
Equation: v_ij = [h1i*h1j, h1i*h2j + h2i*h1j, h2i*h2j,
h3i*h1j + h1i*h3j, h3i*h2j + h2i*h3j, h3i*h3j]
"""
h_i = H[:, i]
h_j = H[:, j]
v = np.array(
[
h_i[0] * h_j[0],
h_i[0] * h_j[1] + h_i[1] * h_j[0],
h_i[1] * h_j[1],
h_i[2] * h_j[0] + h_i[0] * h_j[2],
h_i[2] * h_j[1] + h_i[1] * h_j[2],
h_i[2] * h_j[2],
]
)
return v
def solve_intrinsic_matrix(homographies):
"""
Solves for K given a list of Homography matrices (at least 3).
Uses Zhang's constraints to solve Gb = 0.
"""
# 1. Build Matrix G (2N x 6)
G = []
for H in homographies:
# Constraint 1: h1^T B h2 = 0 => v12^T b = 0
v12 = get_v_ij(H, 0, 1)
# Constraint 2: h1^T B h1 - h2^T B h2 = 0 => (v11 - v22)^T b = 0
v11 = get_v_ij(H, 0, 0)
v22 = get_v_ij(H, 1, 1)
v_diff = v11 - v22
G.append(v12)
G.append(v_diff)
G = np.array(G)
# 2. Solve Gb = 0 using SVD
# b is the eigenvector corresponding to smallest eigenvalue
U, S, Vt = np.linalg.svd(G)
b = Vt[-1]
# 3. Reconstruct B matrix
# b = [B11, B12, B22, B13, B23, B33]
B = np.array([[b[0], b[1], b[3]], [b[1], b[2], b[4]], [b[3], b[4], b[5]]])
# Check if B is positive definite. If not, flip sign of b (scale ambiguity).
# B must be positive definite for Cholesky.
# Simple check: B[0,0] should be positive (related to 1/fx^2)
if B[0, 0] < 0:
B = -B
# 4. Cholesky Decomposition: B = L L^T
try:
L = np.linalg.cholesky(B)
except np.linalg.LinAlgError:
print(
"Error: B is not positive definite. Data too noisy or singular configuration."
)
return np.eye(3)
# 5. Compute K
# B = (K^-1)^T (K^-1) = L L^T => L = (K^-1)^T => L^T = K^-1
# K = inv(L^T)
K_inv = L.T
K = np.linalg.inv(K_inv)
# Normalize K so K[2,2] = 1
K = K / K[2, 2]
return K
if __name__ == "__main__":
# Ground Truth K
K_true = np.array([[1000, 0, 320], [0, 1000, 240], [0, 0, 1]])
# Generate random rotations/translations to make fake Homographies
H_list = []
for i in range(3):
# Random rotation (using small angles for simplicity)
ang = np.random.rand(3) * 0.5
from scipy.spatial.transform import Rotation
R = Rotation.from_euler("xyz", ang).as_matrix()
t = np.random.rand(3, 1)
# H = K [r1, r2, t]
# We remove the 3rd column of R (Z-axis)
H_sim = K_true @ np.hstack((R[:, [0, 1]], t))
H_sim = H_sim / H_sim[2, 2] # Normalize
H_list.append(H_sim)
# Solve
K_est = solve_intrinsic_matrix(H_list)
print("True K:\n", K_true)
print("\nEstimated K:\n", np.round(K_est, 2))